Reposted from the original at https://blog.stephenturner.us/p/bluesky-analysis-claude-llama-tidyverse.
Summary, translation, and sentiment analysis of Bluesky posts on a topic using atrrr, tidyverse/ellmer, and mlverse/mall with Claude and other open models, and gistr to put the summary on GitHub.
...
Background
Bluesky, atrrr, local LLMs
I’ve written a few posts lately about Bluesky — first, Bluesky for Science, about Bluesky as a home for Science Twitter expats after the mass eXodus, another on using the atrrr package to expand your Bluesky network. I’ve also spent some time looking at R packages to provide an interface to Ollama. I wrote about this generically last summer, and quickly followed that up with a post about an R package that uses Llama 3.1 for summarizing bioRxiv and medRxiv preprints.
Using frontier models in R
In those posts above I’m using a local LLM (some variant of the llama model). Meta claims that the newest llama3.3-70B model achieves performance on par with GPT-4 and the much larger llama3.1-405B. It’s good (really good for an open-ish model), but I don’t agree with this claim (and neither does the Chatbot Arena leaderboard).
Enter ellmer (ellmer.tidyverse.org) — a recent addition to the tidyverse that provides interfaces to frontier models like OpenAI’s ChatGPT, Anthropic’s Claude, Google’s Gemini, and others (along with local models via Ollama).
I bought a few bucks worth of Claude API credits to see how Claude would compare to some of these open source models for a simple summarization task.
The task
Bluesky for science has really taken off since X’s new management has let the place degrade into the sewer that it is. It’s such a lively place and if I go a day or two without scrolling through my feeds I’ll miss a lot of what’s happening in the R community and other interest groups.
I subscribe to the AI News newsletter, where every day I get an AI-summarized digest of what’s happening in the AI world across Twitter, Reddit, various Discords, and other forums (fora?). I was curious if I could do something similar with the #Rstats hashtag on Bluesky.
The setup
Here’s the basic setup. Here’s the code on GitHub.
Use the atrrr package to retrieve 1000 Bluesky posts tagged with #Rstats. I typically see about 100 posts per day, so 1000 is enough to capture everything in the past week.
Use dplyr to do some cleaning: Limit posts to just the previous 7 days, create a link to the post URL from the at:// URI, arrange descending by the most liked posts, having at least 5 likes, extract the post text to feed to the LLM, and extract a bullet point list of post text for the top 10 posts to include in the summary.
Use the ellmer package to summarize these posts using Claude 3.5 Sonnet and various open models through Ollama. Run three iterations of each, and choose the best result out of the three (subjective).
Claude 3.5 Sonnet
Llama3.3
Gemma2
Mistral
Use ROpenSci’s gistr package to programmatically post the resulting markdown as a GitHub gist.
The system prompt I’m using for each of these is:
What are the top posts on #rstats Bluesky last week? I will provide you with data showing the author, post URL, and post text from the top 100 most liked posts tagged with #Rstats. Remember, this is bluesky, and posts are called posts, NOT tweets. Make sure you call them posts, not tweets. I want you to return a summary of the top posts, in markdown. Who posted about what? And why is it important? Give examples. Give me a list of topics, a list of common hashtags, and author names. Also talk about the general overall tone or sentiment. You can link to a specific post by using the URL provided, where the link text is the author's name, like '[author](url) posted <text>'. Do not include any preample or postscript text or followup question suggestions. Here are the posts.
Results
Note: I ran the code linked above to generate these results for the week of December 18-24, 2024. You’ll get different results running the same code today.
The result from Claude 3.5 Sonnet was far and above the best summary, and it followed my system prompt instructions better than any of the open source models. After checking my usage, this query cost about $0.01-$0.02 cents.
Claude 3.5 Sonnet
Results from Claude 3.5 Sonnet:
Summary
Produced with claude-3-5-sonnet-20241022
Here's a summary of the top #rstats posts on Bluesky from the past week:
Key Announcements and News:
Posit (formerly RStudio) joined the Open Source Pledge, committing $2000/year per developer to support open source software
The RStudio IDE and Posit Workbench received a major update with Positron Pro support
useR 2025 will be held at Duke University in August 2024
Several notable package releases including rms 6.9-0, {chopin} for spatial analysis, and {rayrender} v0.38.0
Popular Topics and Themes:
Package Development & Updates
Data Visualization
Tutorials & Learning Resources
Statistical Methods
Mobile Development
Generative Art
Notable Tutorial Content:
Jarrett Byrnes shared a tutorial on Bayesian SEM using brms
Danielle Navarro wrote about regular expressions in R
Multiple posts about the easystats ecosystem for Bayesian analysis
Innovation & New Tools:
Colin announced Rlinguo, a mobile app for running R on phones
New developments in web-based R applications using webR and Shinylive
Various updates to visualization packages and tools
Community Engagement:
Active TidyTuesday participation analyzing D&D spell data
Significant discussion around generative art and visualization
Strong focus on teaching and learning resources
The overall tone of the R community on Bluesky is overwhelmingly positive and collaborative, with a mix of technical discussion, creative applications, and mutual support. There's a strong emphasis on sharing knowledge and resources, particularly around data visualization and statistical methods.
Common Hashtags: #rstats #dataviz #tidytuesday #pydata #statistics #dataBS #rtistry #bayesian #shiny #ggplot2
Notable Authors:
Hadley Wickham
Danielle Navarro
Frank Harrell
Tyler Morgan-Wall
Sharon Machlis
Colin Fay
Nicola Rennie
The community appears to be particularly excited about new developments in mobile and web-based R applications, as well as advances in statistical computing and visualization tools. There's also a strong strain of creativity running through the posts, with many sharing artistic applications of R programming.
Top posts
Top 10 posts:
Hadley Wickham: Very proud to announce that @posit.co has joined the opensourcepledge.com. We're committing to spending $2000 / developer / year to support open source software that we use (but don't develop): posit.co/blog/posit-p... #rstats #pydata
Jarrett Byrnes: OK, here is a very rough draft of a tutorial for #Bayesian #SEM using #brms for #rstats. It needs work, polish, has a lot of questions in it, and I need to add a references section. But, I think a lot of folk will find this useful, so.... jebyrnes.github.io/bayesian_sem... (use issues for comments!)
Danielle Navarro: Some words on regular expressions in #rstats. In which I finally wrap my head around the fact that there are at least three different regex engines in widespread usage in R, they support different features, and they all hate me blog.djnavarro.net/posts/2024-1...
Colin 🤘🌱🏃♀️: #RStats I'm so, so thrilled to finally share the release of Rlinguo, a mobile app that runs R 📱 This is a fully, native mobile app that uses #webR in the backend to run R functions. Available on the App Store (iPhone) & Play Store (Android) rtask.thinkr.fr/introducing-...
Erik Reinbergs: These bayes tutorials are the first ones i've actually understood. Looking forward to the rest of the being finished. Go @easystats.bsky.social teams! easystats.github.io/bayestestR/i... #rstats
Dave H: Just pushed a Christmas update to {ggblanket}. I decided to support colour blending. It uses {ggblend} under the hood (thanks @mjskay.com), which uses graphics features developed by Paul Murrell. Give {ggblanket} and {ggblend} a star, if you find them useful. Oh, and have a merry Christmas #rstats
Owen Phillips: I created a thing that puts every player's box score stats from the night before in one interactive table. Ive been using it every morning to quickly check who had a good/bad game instead of looking at 10 different box scores updates nightly #rstats #gt #quarto thef5.quarto.pub/boy/
Hadley Wickham: I discovered that the original plyr website still exists: had.co.nz/plyr/. It's hard to imagine that this used to be a better than average package website #rstats
Frank Harrell: 1/2 Major release 6.9-0 of #RStats rms package now available on CRAN w/complete re-write of the binary/ordinal logistic regression function lrm. lrm began in 1980 as SAS PROC LOGIST so it was time for a re-do for iterative calculations. Details are at fharrell.com/post/mle
Joe Chou: As far as I’m concerned, #webr and #shinylive is indistinguishable from magic. I can’t believe that since first installing #quarto-webr a few hours ago till now, I’ve gotten a relatively complicated #shinyapp exported, deployed, and running serverless. This is magic. #rstats
Open-source models: Llama3.3, Gemma2, Mistral
I’m not showing the other results here — the links go to a Markdown file as a GitHub gist showing results from running the same prompt with Llama3.3, Gemma2, and Mistral. They’re very different! Gemma just provided a very high-level overview that could have come from any collection of posts about R, and mistral just listed out individual summaries of each post instead of summarizing the whole collection. Llama3.3 might have been a bit better, but overall, it seems that the open source models aren’t following my system prompt as well as Claude 3.5 Sonnet is.
Bonus: sentiment, summary, and translation
While the ellmer package demonstrated above provides a straighforward way to prompt local or frontier models with text, the mall package in the mlverse (mlverse.github.io/mall) provides functions for running LLM predictions against a data frame. It’s API is similar in the R and Python packages.
If I have the data frame created above with the post text in d$text, I can run a sentiment analysis, create a 5 word summary, and translate into Spanish with the code below. New columns are added to the data, prefixed by a dot.
d |>
llm_sentiment(text) |>
llm_summarize(text, max_words = 5) |>
llm_translate(text, "spanish")Here’s what the top 10 most liked posts from the past week are, summarized in 5 words, translated into Spanish:
Hadley Wickham (posit joins opensourcepledge): Estamos muy orgullosos de anunciar que @posit.co ha unido a opensourcepledge.com. Estamos comprometidos a gastar $2000 / desarrollador / año para apoyar software de código abierto que usamos (pero no desarrollamos): posit.co/blog/posit-p... #rstats #pydata
Jarrett Byrnes (tutorial on bayesian sem rstats): Este es un tutorial muy desgloseado para el uso de SEM bayesiano utilizando brms en RStats. Necesita ser revisado, pulido y tener muchas preguntas resueltas, pero creo que muchos personas lo encontrarán útil, así que... jebyrnes.github.io/bayesian_sem... (usen los problemas para hacer comentarios!)
Danielle Navarro (three r regex engines exist): Hay al menos tres diferentes motores de expresiones regulares en uso amplio en R, que apoyan diferentes características y todos me odian.
Colin 🤘🌱🏃♀️ (mobile app for r programming): Estoy extremadamente emocionado de compartir finalmente el lanzamiento de Rlinguo, una aplicación móvil que ejecuta R.
Erik Reinbergs (bayes tutorials finally make sense): Estos tutoriales de Bayes son los primeros que he entendido. Estoy emocionado de ver el resto terminado. ¡Buena suerte a todos en @easystats.bsky.social!
Dave H (pushed christmas update to ggblanket): Acabo de lanzar una actualización de Navidad para {ggblanket}. Decidí apoyar la mezcla de colores. Utiliza {ggblend} detrás del escenario (gracias @mjskay.com), que utiliza características gráficas desarrolladas por Paul Murrell. Dale un estrella a {ggblanket} y {ggblend}, si los encuentras útiles. ¡Que tengas un feliz Navidad #rstats
Owen Phillips (interactive box score table): Crea una cosa que agrega todos los estadísticas de puntuación del tablero de cada jugador de la noche anterior en una tabla interactiva. He estado utilizandola cada mañana para revisar rápidamente quién tuvo un buen/buen juego en lugar de mirar las actualizaciones de diez diferentes partidos nocturnos #rstats #gt #quarto thef5.quarto.pub/boy
Hadley Wickham (original plyr website still exists): Descubrí que la página web original de plyr aún existe: had.co.nz/plyr/. Es difícil imaginar que esto era una página de paquete mejor que promedio. #rstats
Frank Harrell (rms package version released): La versión 6.9-0 del paquete #RStats rms ahora está disponible en CRAN con una completa revisión de la función de regresión logística binomial lrm, que comenzó en 1980 como PROC LOGIST en SAS y requería una reescritura para cálculos iterativos.
Joe Chou (webr and shiny are magic): En mi opinión, #webr y #shinylive son indistinguibles del magia. No puedo creer que desde que instale #quarto-webr unas horas atrás hasta ahora, he logrado exportar, desplegar y correr una aplicación #shinyapp relativamente complicada serverless. Esto es magia. #rstats
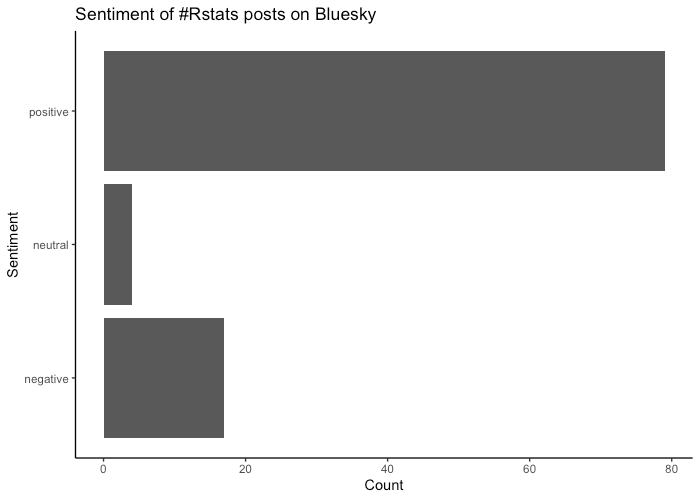
Here’s a count of the sentiment classification for each post.
I was curious what the negative sentiment posts were. The most liked negative sentiment post was Danielle Navarro’s blog post on different regex engines in R and the difficulty using them consistently. Others had a degree cynicism that the model picked up on.
"Some words on regular expressions in #rstats. In which I finally wrap my head around the fact that there are at least three different regex engines in widespread usage in R, they support different features, and they all hate me blog.djnavarro.net/posts/2024-1..."
"Found a bug in my code and after fixing it, it stopped working. Wonderful Christmas gift :3 #rstats"
"Turning a 10-minute #rstats project into a two-hour one because I refuse to break my pipe and save an intermediate object to memory."